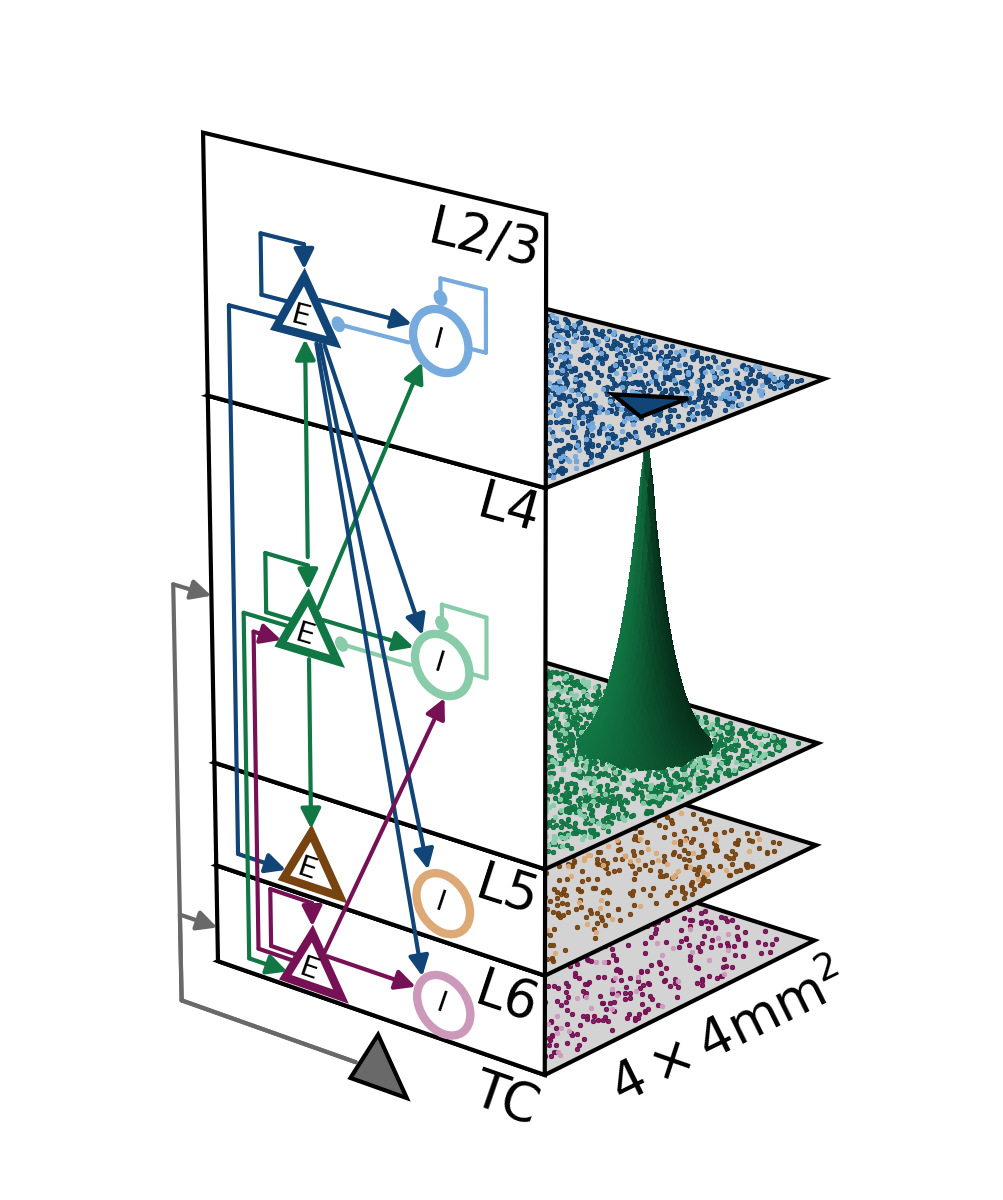
Overview¶
The mesocircuit framework allows for simulating and analyzing spiking cortical network models similar to the microcircuit by Potjans and Diesmann[1]. Subsequent predictions of the local field potential can be obtained with the method described by Hagen et al.[2]. The mesocircuit model extends the previous works with spatial structure, including local distance-dependent connectivity.
Structure of the repository¶
scripts: Examples demonstrating how to use the framework (see Examples below).mesocircuit: Mesocircuit Python package.mesocircuit_framework.py: Parameterspace evaluation and definition of the main classesMesocircuitExperimentandMesocircuit.parameterization: Base parameters of the spiking model distinguishing between system (machine), simulation, network, analysis, and plotting parameters. Tools for deriving dependent parameters.simulation: Network class of the spiking model, extending the PyNEST example Cortical microcircuit model.analysis: Preprocessing and statistical analysis of spike data.plotting: Stand-alone plotting functions and figure definitions.lfp: Morphologies and other scripts related to local field potentials.helpers: Generic helper functions, e.g., handling of input-output and MPI parallelism.run: Concrete steps of job execution for network simulation, analysis, and plotting (both for the spiking model and the local field potential).
tests: Test suite with unit tests.docs: Source code of the documentation.
Installation¶
The main model requires high-performance computing and we provide installation instructions for the supercomputer JURECA. For local testing on a laptop, the mesocircuit can be set up via conda.
Examples¶
Getting started¶
To familiarize yourself with the general concepts of the mesocircuit model, you may first check out the Jupyter notebook simulating only one layer of the mesocircuit model set up as a tutorial, see scripts/reduced_mesocircuit.
Run mesocircuit¶
This main example script allows you to run the full mesocircuit.
Per default this is an upscaled version of area V1 of the multi-area model of macaque monkey cortex (Schmidt et al.[3], Schmidt et al.[4]).
You can easily change the parameterization to a different model using the predifined configurations in parametersets.py.
While the mesocircuit model requires high-performance computing, you can test out a downscaled version locally on your laptop.
Parameter space exploration¶
If you are interested in testing parameter ranges, here is an example for spanning a two-dimensional parameter space. The framework automatically runs the simulation and analysis for all parameter combinations and generates comparison figures.
Mean-field theory¶
The framework integrates mean-field predictions (e.g., firing rates, power spectra) using NNMT - Neuronal Network Meanfield Toolbox (Layer et al.[5]). Note, however, that a good agreement between theory and simulation can be achieved for models such as the cortical microcircuit by Potjans and Diesmann[1], but for spatially structured models discrepancies are expected as the toolbox does not include the corresponding functionality yet.
Network sketches¶
Network diagrams of a reference network model (without space) and the upscaled spatially structured network model.
Manuscript figures¶
These files allow the reproduction of the figures in the manuscript accompanying this repository (currently this manuscript is being finalized).